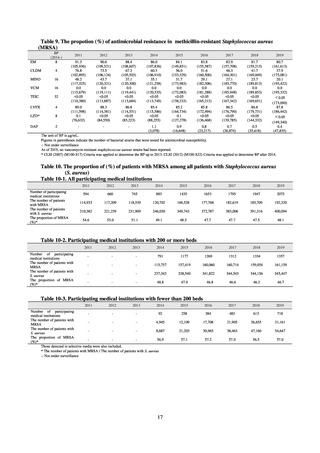
【参考資料3】【英版R4.1.17】Nippon AMR One Health Report (NAOR) 2020 (93 ページ)
出典
| 公開元URL | https://www.mhlw.go.jp/stf/newpage_23261.html |
| 出典情報 | 国際的に脅威となる感染症対策関係閣僚会議 薬剤耐性ワンヘルス動向調査検討会(第9回 1/17)《厚生労働省》 |
ページ画像
プレーンテキスト
資料テキストはコンピュータによる自動処理で生成されており、完全に資料と一致しない場合があります。
テキストをコピーしてご利用いただく際は資料と付け合わせてご確認ください。
1) Overview
JVARM is a nationwide system for monitoring antimicrobial-resistant bacteria among animals. This monitoring has
been conducted by the Ministry of Agriculture, Forestry and Fisheries since 1999 through its network of connections with
livestock hygiene service centers across Japan. JVARM provides globally important information and is cited as an example
of a monitoring system in the WHO report “Antimicrobial resistance: global report on surveillance 2014.”
Under JVARM, three types of monitoring are conducted: (1) monitoring of the volumes of use of antimicrobials
(estimated from the volumes of sales); (2) monitoring of antimicrobial resistance among indicator bacteria and foodborne
pathogens derived from healthy animals; and (3) monitoring of antimicrobial resistance in pathogenic bacteria (clinical
isolates) derived from diseased animals. While verifying the efficacy of veterinary antimicrobials, JVARM also provides
basic data for risk assessment and risk management concerning antimicrobial resistance, taking into account influence on
human healthcare (Figures 1). The results of JVARM are published on the website of the National Veterinary Assay
Laboratory, Ministry of Agriculture, Forestry and Fisheries [2]. In FY2016, reviews were carried out to consider how to
strengthen antimicrobial resistance surveillance in aquatic animals and how to conduct antimicrobial resistance surveillance
in companion animals, in accordance with the strategies of the National Action Plan on AMR. Antimicrobial resistance
surveillance in diseased dogs and cats was launched in FY2017 and in healthy dogs and cats in FY2018.
Figure 1. Overview of veterinary antimicrobial resistance monitoring
2) System for the antimicrobial resistance monitoring
When JVARM first began, surveillance of foodborne pathogenic bacteria and indicator bacteria from healthy animals
was carried out using samples of strains isolated and identified from the feces of food-producing animals collected at farms
by livestock hygiene service centers. Surveillance using strains isolated and identified by the contracted testing agency
from feces collected at animal and poultry slaughterhouses was launched in FY2012, as this facilitated more intensive
sampling at a stage closer to the final food product. In FY2016, as it had been confirmed that there was no major difference
in the findings of both surveys, JVARM shifted completely from sampling at farms to sampling at animal and poultry
slaughterhouses (Figure 2). Bacteria were isolated from feces samples using species-selective media and data are based on
one strain per bacterial species per farm (the farm’s representative strain).
In the case of clinical isolates from food-producing animals, bacterial strains isolated and identified from materials for
pathological appraisal by livestock hygiene service centers across the country were collected. One or two strains isolated
from a different individual affected in a single case of infectious disease were collected for the monitoring. The MIC values
for these strains are measured by the National Veterinary Assay Laboratory using a broth microdilution method based on
the CLSI Criteria (Figure 3). The scope of antimicrobial monitoring includes a broad range of active ingredients that are
considered important in antimicrobials used exclusively for animals, antimicrobials used for both animals and humans, and
antimicrobial feed additives, among others. Antimicrobial agents subject to monitoring are selected for each bacterial
species, according to the past monitoring results and Chapter 6.7 of the OIE Terrestrial Animal Health Code.[3]
The framework for surveillance of companion animals was determinedbased on the results of deliberations by the
Working Group for the Surveillance of AMR in Companion Animals. In 2017, bacterial strains isolated from diseased dogs
and cats began to be collected from clinical laboratories (Figure 4). Since 2018, samples from healthy dogs and cats have
been collected from veterinary clinics across the country with the cooperation of the Japan Veterinary Medical Association
(Figure 5). All bacteria were isolated from samples using species-selective media, and adopted one strain per bacterial
92
関連画像
ページ内で利用されている画像ファイルです。